Accepts a formula to run an xgboost model. Automatically determines whether the formula is for classification or regression. Returns the xgboost model.
tidy_xgboost(
.data,
formula,
...,
mtry = 0.75,
trees = 500L,
min_n = 2L,
tree_depth = 7L,
learn_rate = 0.05,
loss_reduction = 1,
sample_size = 0.75,
stop_iter = 15L,
counts = FALSE,
tree_method = c("auto", "exact", "approx", "hist", "gpu_hist"),
monotone_constraints = 0L,
num_parallel_tree = 1L,
lambda = 0.5,
alpha = 0.1,
scale_pos_weight = 1,
verbosity = 0L,
validate = TRUE,
booster = c("gbtree", "gblinear")
)Arguments
- .data
dataframe
- formula
formula
- ...
additional parameters to be passed to
set_engine- mtry
# Randomly Selected Predictors; defaults to .75; (xgboost: colsample_bynode) (type: numeric, range 0 - 1) (or type: integer if
count = TRUE)- trees
# Trees (xgboost: nrounds) (type: integer, default: 500L)
- min_n
Minimal Node Size (xgboost: min_child_weight) (type: integer, default: 2L); [typical range: 2-10] Keep small value for highly imbalanced class data where leaf nodes can have smaller size groups. Otherwise increase size to prevent overfitting outliers.
- tree_depth
Tree Depth (xgboost: max_depth) (type: integer, default: 7L); Typical values: 3-10
- learn_rate
Learning Rate (xgboost: eta) (type: double, default: 0.05); Typical values: 0.01-0.3
- loss_reduction
Minimum Loss Reduction (xgboost: gamma) (type: double, default: 1.0); range: 0 to Inf; typical value: 0 - 20 assuming low-mid tree depth
- sample_size
Proportion Observations Sampled (xgboost: subsample) (type: double, default: .75); Typical values: 0.5 - 1
- stop_iter
# Iterations Before Stopping (xgboost: early_stop) (type: integer, default: 15L) only enabled if validation set is provided
- counts
if
TRUEspecifymtryas an integer number of cols. DefaultFALSEto specifymtryas fraction of cols from 0 to 1- tree_method
xgboost tree_method. default is
auto. reference: tree method docs- monotone_constraints
an integer vector with length of the predictor cols, of
-1, 1, 0corresponding to decreasing, increasing, and no constraint respectively for the index of the predictor col. reference: monotonicity docs.- num_parallel_tree
should be set to the size of the forest being trained. default 1L
- lambda
[default=.5] L2 regularization term on weights. Increasing this value will make model more conservative.
- alpha
[default=.1] L1 regularization term on weights. Increasing this value will make model more conservative.
- scale_pos_weight
[default=1] Control the balance of positive and negative weights, useful for unbalanced classes. if set to TRUE, calculates sum(negative instances) / sum(positive instances). If first level is majority class, use values < 1, otherwise normally values >1 are used to balance the class distribution.
- verbosity
[default=1] Verbosity of printing messages. Valid values are 0 (silent), 1 (warning), 2 (info), 3 (debug).
- validate
default TRUE. report accuracy metrics on a validation set.
- booster
defaults to 'gbtree' for tree boosting but can be set to 'gblinear'
Value
xgb.Booster model
Details
In binary classification the target variable must be a factor with the first level set to the event of interest. A higher probability will predict the first level.
reference for parameters: xgboost docs
Examples
options(rlang_trace_top_env = rlang::current_env())
# regression on numeric variable
iris %>%
framecleaner::create_dummies(Species) -> iris_dummy
#> 1 column(s) have become 3 dummy columns
iris_dummy %>%
tidy_formula(target= Petal.Length) -> petal_form
iris_dummy %>%
tidy_xgboost(
petal_form,
trees = 20,
mtry = .5
) -> xg1
#> accuracy tested on a validation set
#> # A tibble: 3 × 2
#> .metric .estimate
#> <chr> <dbl>
#> 1 ccc 0.840
#> 2 rmse 0.816
#> 3 rsq 0.945
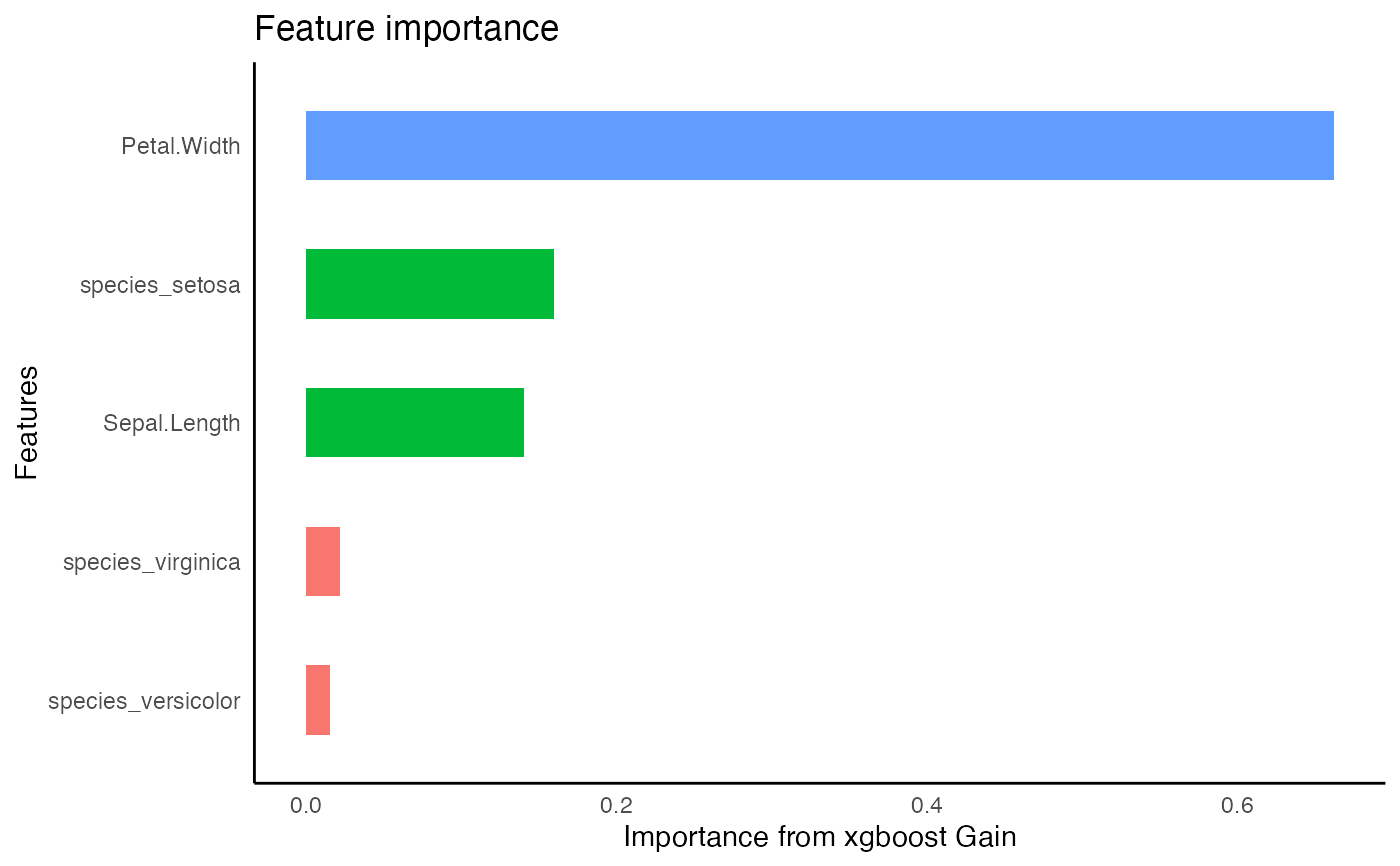 xg1 %>%
tidy_predict(newdata = iris_dummy, form = petal_form) -> iris_preds
#> created the following column: Petal.Length_preds_xg1
iris_preds %>%
eval_preds()
#> # A tibble: 3 × 5
#> .metric .estimator .estimate model target
#> <chr> <chr> <dbl> <chr> <chr>
#> 1 ccc standard 0.884 xg1 Petal.Length
#> 2 rmse standard 0.703 xg1 Petal.Length
#> 3 rsq standard 0.980 xg1 Petal.Length
xg1 %>%
tidy_predict(newdata = iris_dummy, form = petal_form) -> iris_preds
#> created the following column: Petal.Length_preds_xg1
iris_preds %>%
eval_preds()
#> # A tibble: 3 × 5
#> .metric .estimator .estimate model target
#> <chr> <chr> <dbl> <chr> <chr>
#> 1 ccc standard 0.884 xg1 Petal.Length
#> 2 rmse standard 0.703 xg1 Petal.Length
#> 3 rsq standard 0.980 xg1 Petal.Length